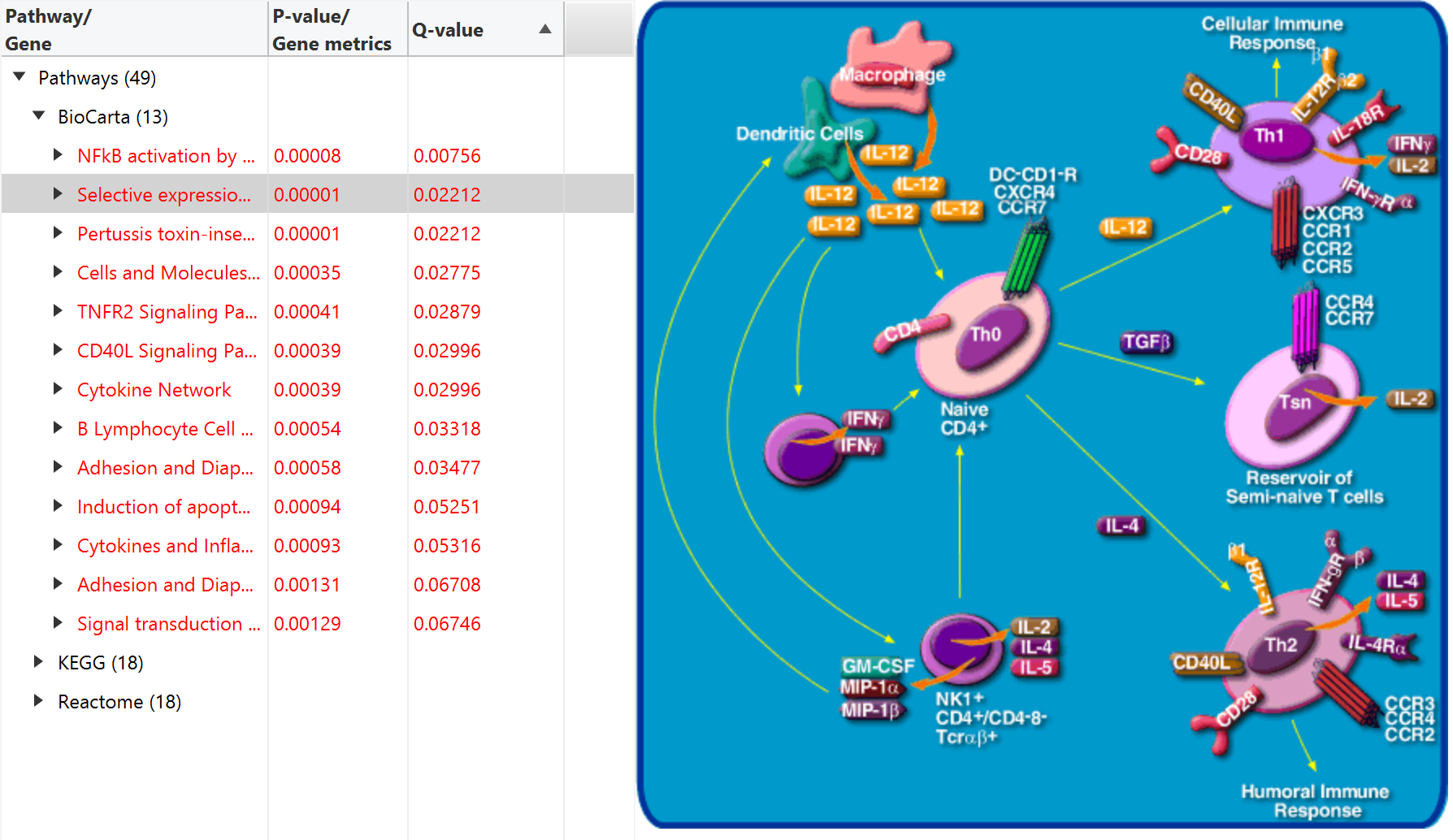
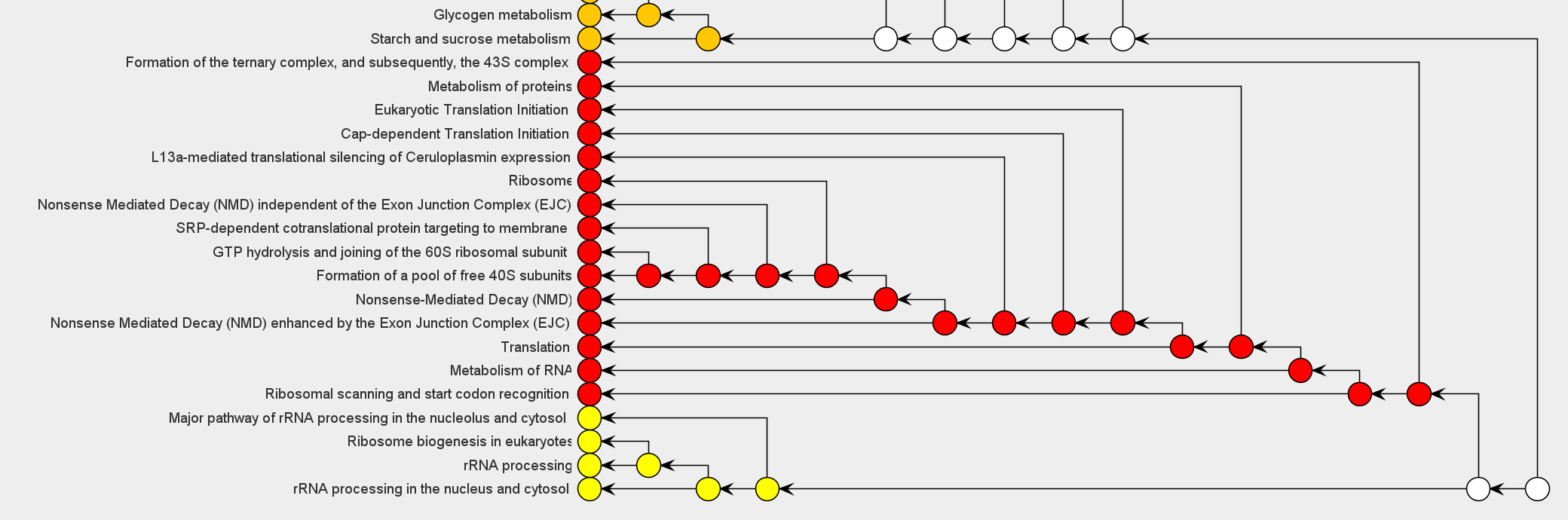
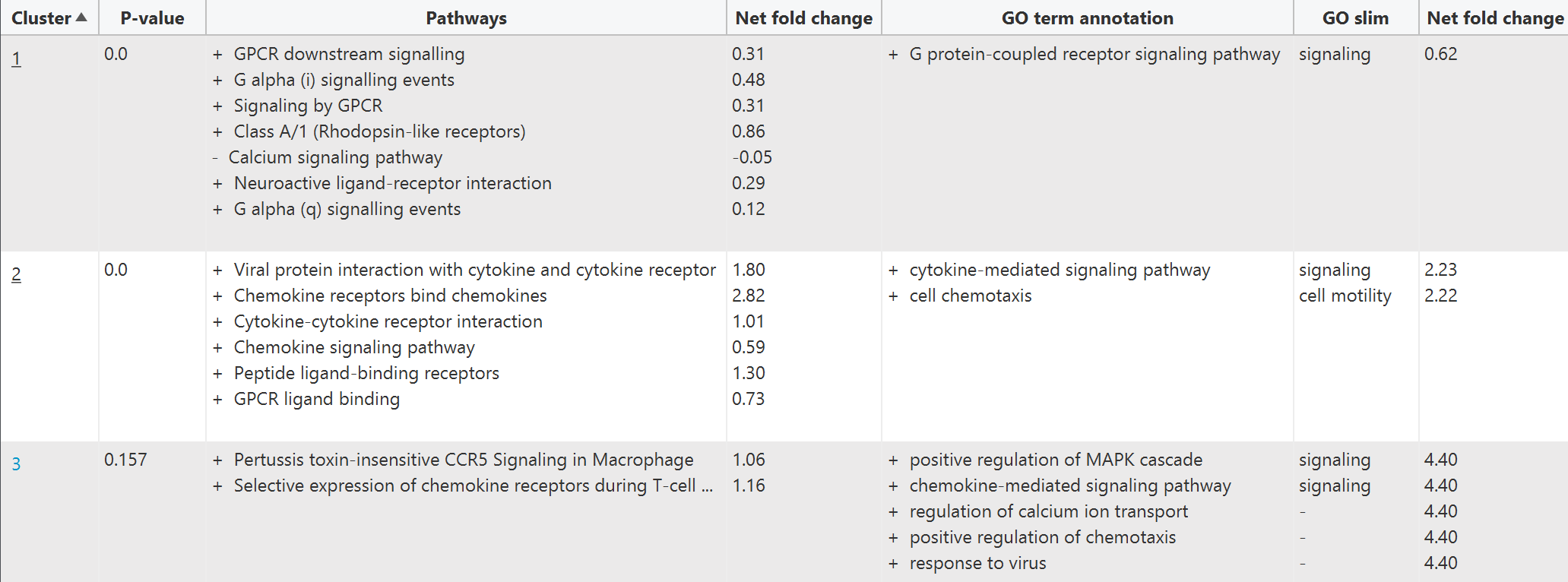
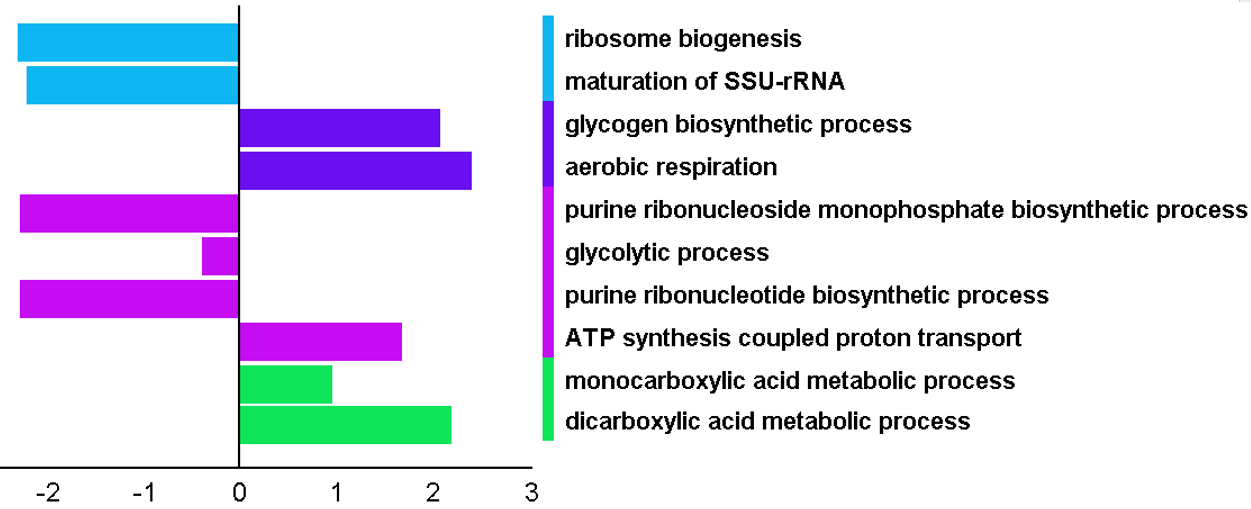
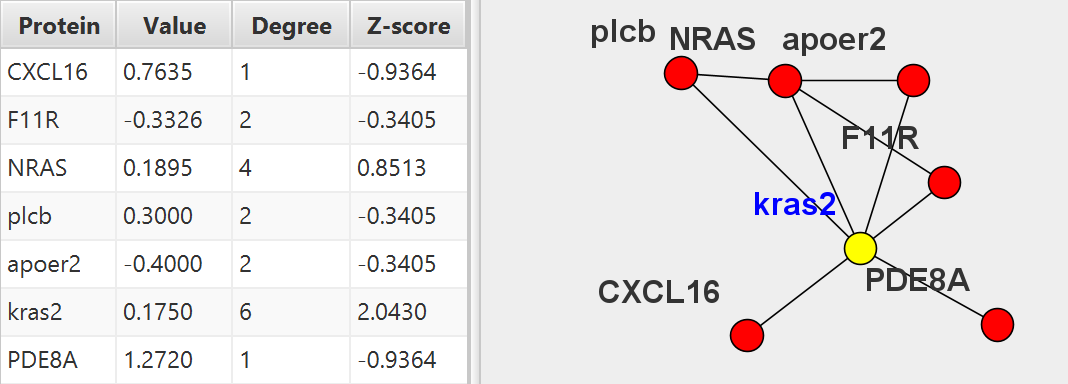
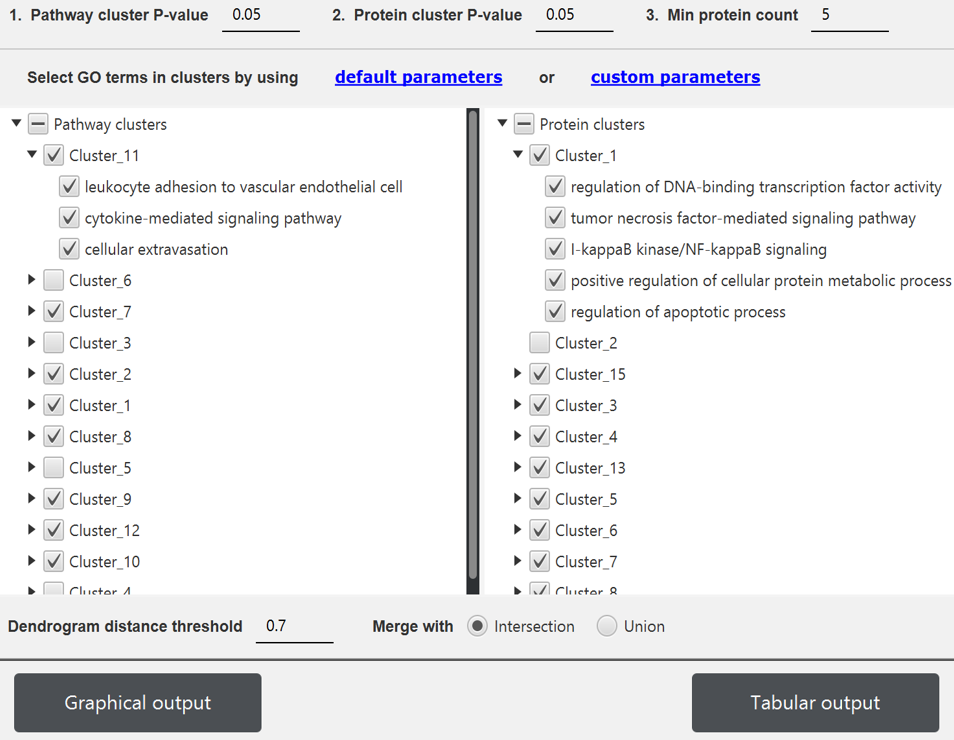
PECPI-GO contains five-step machine-learning methods to decipher complicated
relationships among genes, pathways, GO hierarchical structure, and functional annotation in an
automatic manner:
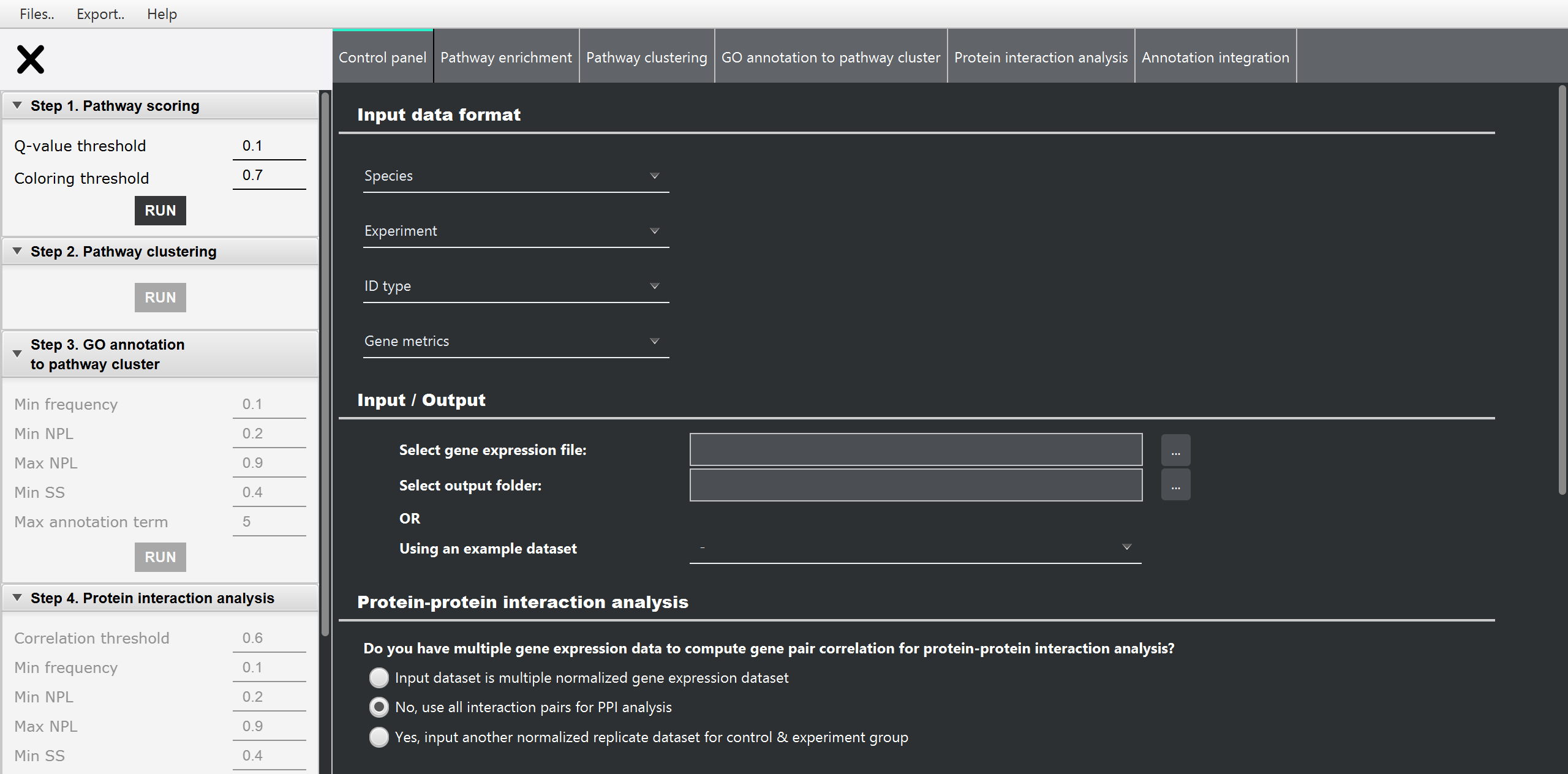
- Pathway enrichment analysis
- Pathway clustering based on graph theory
- GO functional annotation of each pathway cluster
- Protein interaction network analysis
- Integration of transcriptome and interactome analysis